Cryo-Electron Tomography for 3D Volumes
Cryo-Electron Tomography (cryo-ET) provides high-resolution, three-dimensional insights into heterogeneous nanoparticles, including liposomes, lipid nanoparticles (LNPs), exosomes, and more.
Unlike 2D imaging, cryo-ET reconstructs detailed 3D volumes which reveal crucial information about particle morphology and internal composition. This enables a more comprehensive understanding of structural features, heterogeneity, and functional properties.
Cryo-ET Allows Visualization of:
- Distribution of proteins on the surface of nanoparticles
- How encapsulated drugs are arranged inside individual particles
- Particle lamellarity
- Particle interactions

Nanoparticles We Can Perform Cryo-Electron Tomography On

- Extracellular Vesicles & Exosomes
- Extracellular Vesicles & Exosomes
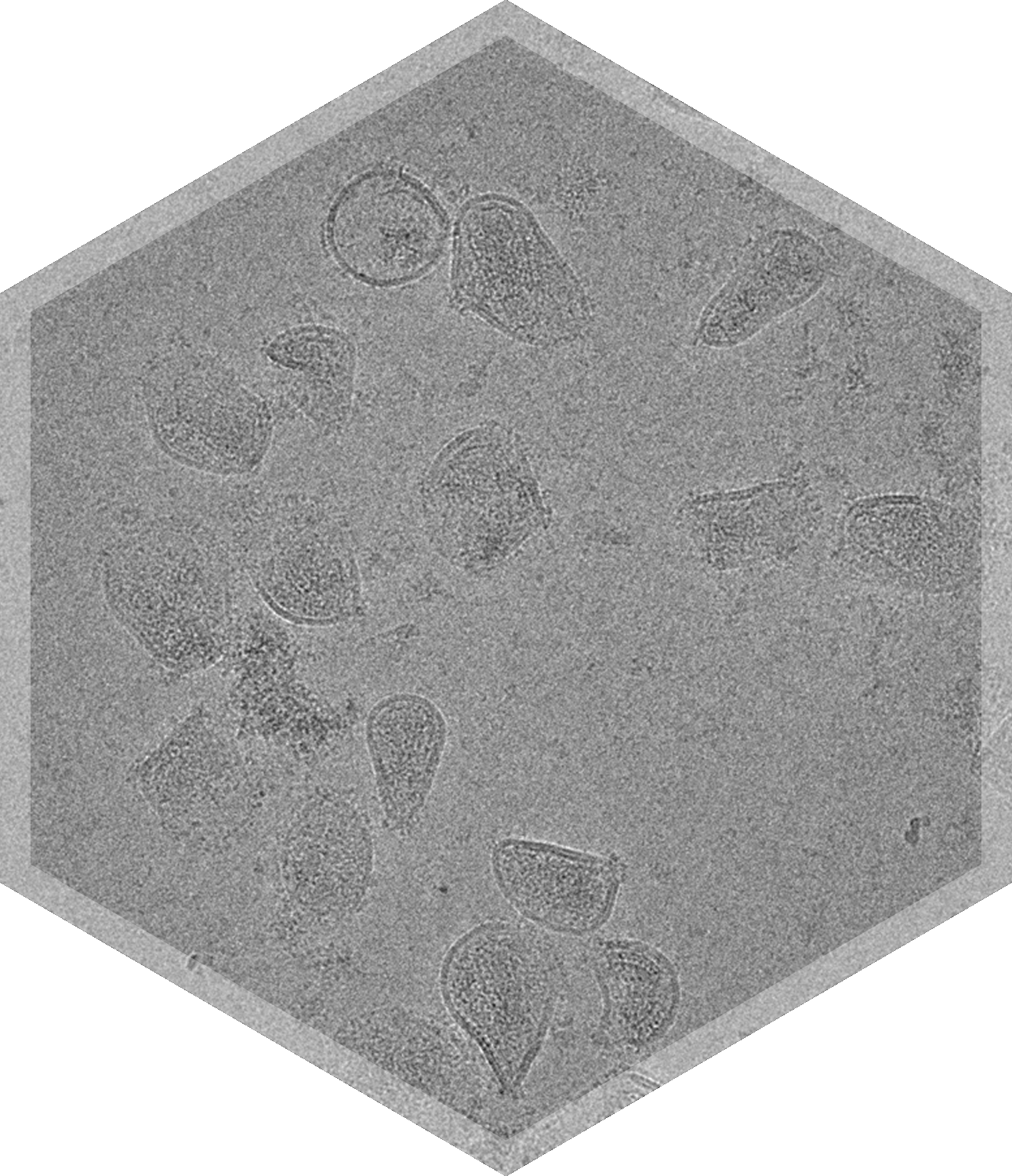
- Lentivirus
- Lentivirus

- Lipid Nanoparticles (LNPs), with mRNA, RNA, DNA payloads
- Lipid Nanoparticles (LNPs), with mRNA, RNA, DNA payloads

- Liposomes
- Liposomes

Cryo-ET allows non-uniform nanoparticles to be interrogated in 3D to assess composition and morphology.
Standard TEM imaging results in 2D projections of 3D objects. This can complicate the interpretation of 3D structure when necessary. While single particle analysis can reveal the 3D structure of small, relatively homogeneous particles, samples consisting of larger and more heterogeneous particles require a different approach. Cryo-Electron tomography (Cryo-ET) is a process for generating a 3D volume from a single field of particles.
A series of 2D projection images is acquired over a range of specimen tilt angles, and these 2D images are used to create a 3D volume of the sample with nanometer resolution. The process is analogous to CT scanning and other medical imaging techniques. The resulting 3D volume can be examined from various orientations and individual slices at different angles through the particles can be analyzed. This can provide a more complete understanding of the particle morphology and internal composition, resolving ambiguities in the 2D projection images.
The analysis of these 3D volumes can inform about the distribution of proteins on the surface of drug delivery vehicles, how encapsulated drugs are arranged inside the drug delivery vehicle, particle lamellarity, as well as particle interactions.










