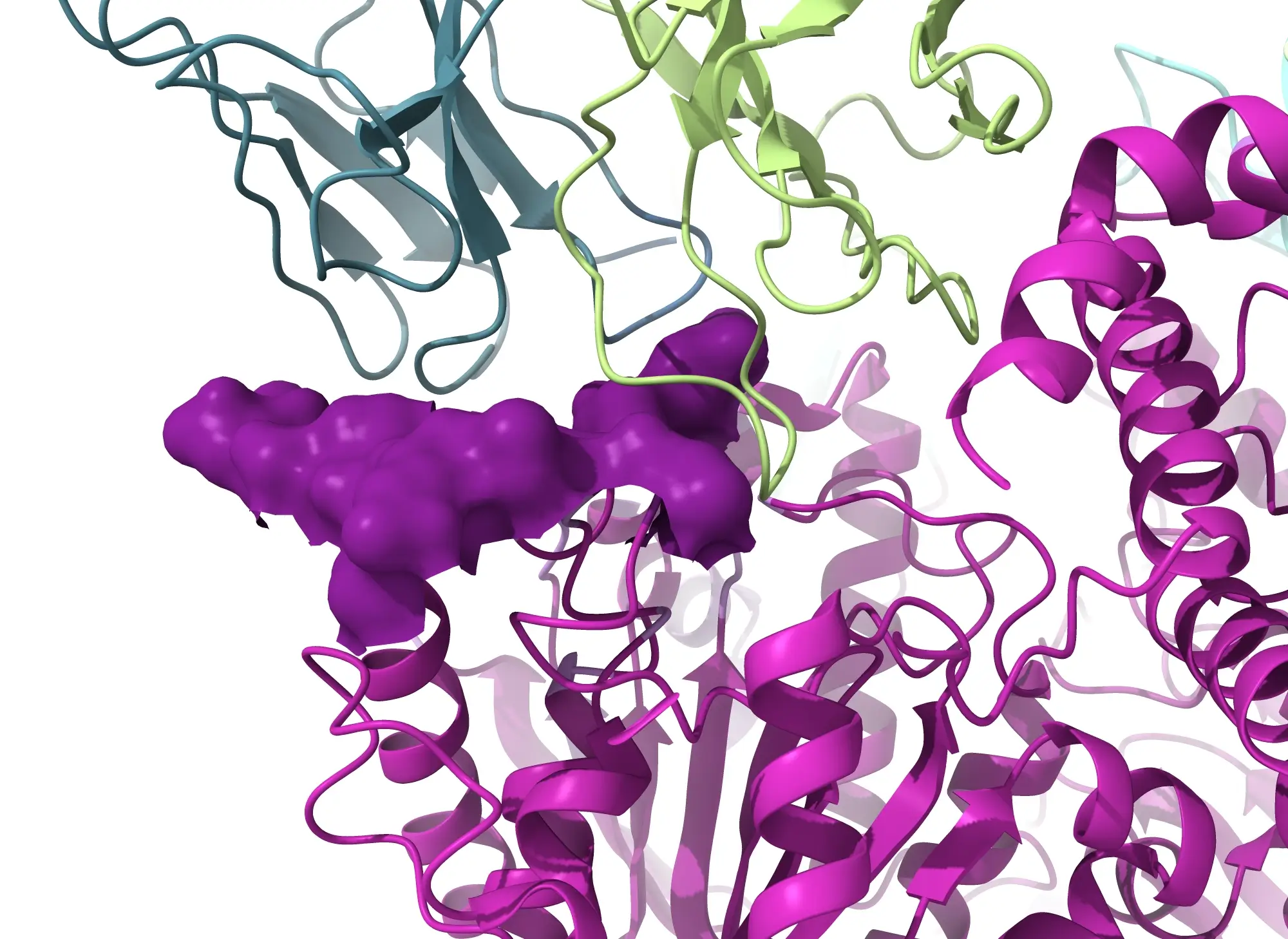
Rapid Epitope Mapping with Cryo-EM
Cryo-EM epitope mapping services can provide a 3D reconstruction to a resolution at the epitope/paratope interface suitable for tracing the chains and assigning 75% of the side chains, within 2 weeks of sample receipt, for suitable samples.
Atomic Level Details Within 2 Weeks
What are the advantages of epitope mapping with cryo-EM?
- Eliminates the need for recombinant Fab production and simplifying the sample production process
- Provides atomic level resolution to visualize the different binding orientations of antibody-antigen complexes
- AB differentiation for patent protection
Cryo-EM epitope mapping can be performed on full length antibodies, Fab fragments in complex with antigens, Fab fragments from a Mab digestion, and antigens complexes with multiple antibodies – no crystallization needed.

Target Classes Solved at NIS
Within 2 Weeks, Researchers Can Obtain Crucial Information About Antibody-Antigen Binding.
Traditionally, scientists have regarded X-ray crystallography (XRC) as the gold standard approach for epitope mapping. While XRC has successfully provided atomic-level details of antibody-antigen complexes, it often encounters significant challenges when dealing with flexible antigens, large complexes, low concentrations, or difficult-to-crystallize samples. This is where cryo-EM becomes important, offering a cutting-edge solution to overcome these limitations.
At NIS, we offer an Epitope Mapping service that uses cryo-EM imaging and data collection to deliver a comprehensive understanding of your antibody-antigen complex. This comprehensive approach enables us to deeply understand the molecular interactions between antibodies and antigens. For suitable samples, we can provide a a 3D reconstruction to a resolution at the epitope/paratope interface suitable for tracing the chains and assigning 75% of the side chains, within 2 weeks of sample receipt. Gain an in-depth understanding of NIS's rapid cryo-EM Epitope Mapping service in our blog here.
An ideal sample is represented by a antigen:FAB complex with a minimum size of 100 kDa, and the epitope should not be located on a loop or other flexible segments of the antigen, and the complex should not exhibit extreme behavior such as severe preferred orientation. If your sample does not fall within these guidelines, we will work closely with you to determine next steps for solving your antibody-antigen complex.
The use of cryo-EM and negative stain TEM imaging for epitope mapping opens up exciting possibilities for unraveling antibody-antigen interactions. These state-of-the-art techniques empower researchers to gain valuable insights into the functional implications of these vital molecular interactions.
Learn more about different techniques for epitope mapping:
Contact us to begin mapping your antibody-antigen complexes.
.webp)