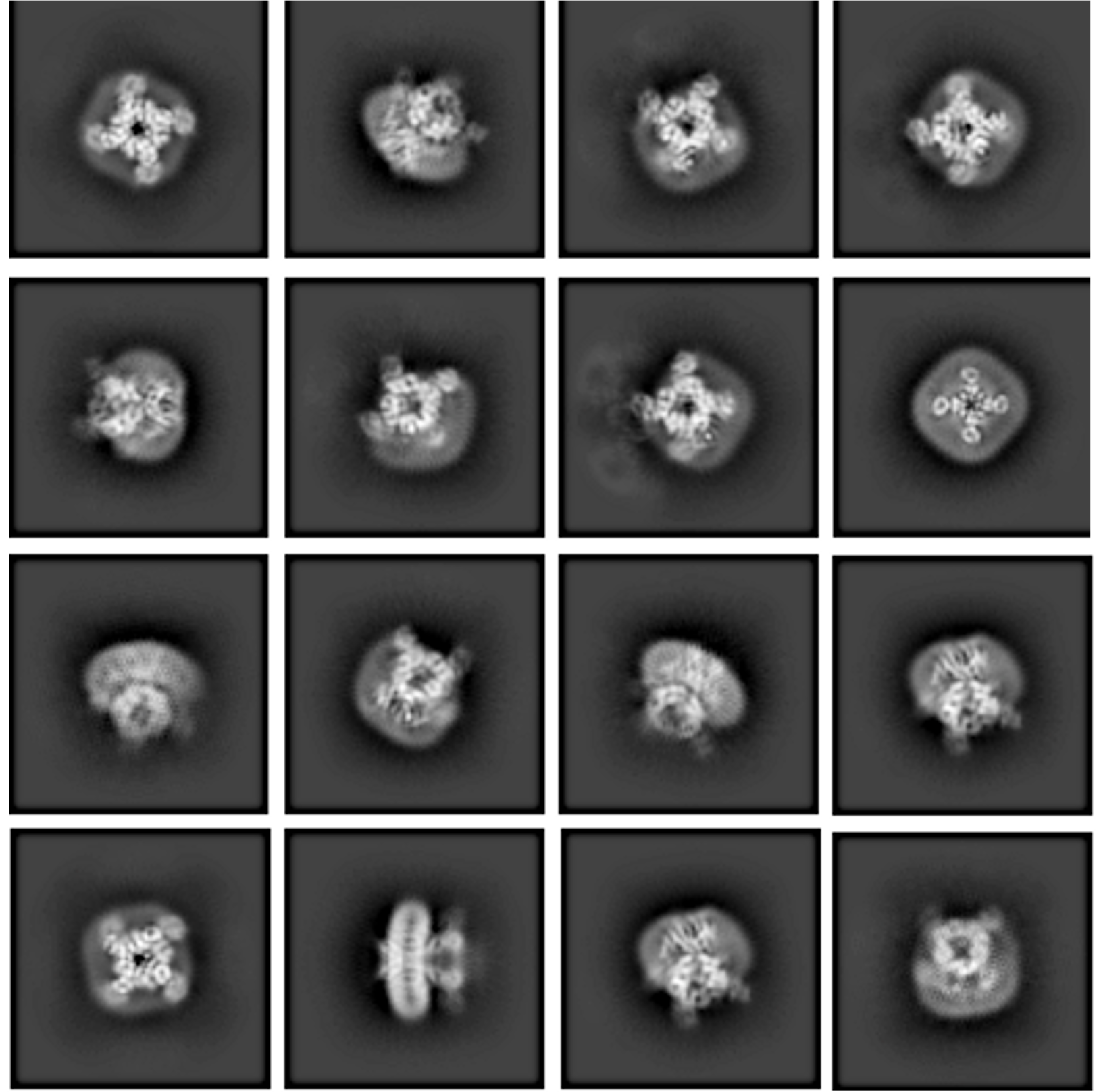
Initial Insights from 2D Class Averages
2D classes are the first step in 3D structure reconstruction. They provide a comprehensive representation of particles in your sample to help determine your next steps forward.
2D class averages can be done in cryo-EM or negative stain TEM imaging.
Gain a First Look at Your Sample with 2D Class Averaging
2D class averages provide a rapid visual insight into:
- Basic particle features, such as aggregation & conformation
- Particle variability
- Preferred orientation
- Domain structures
- Degree of particle flexibility
This information helps assess sample quality for 3D structure reconstruction, improving your chance of success in cryo-EM structure reconstruction.

Target Classes Solved at NIS
- Protein Degraders & Ternary Complexes
- PROTAC® Degraders
- Molecular Glues
- Protein Degraders & Ternary Complexes
- PROTAC® Degraders
- Molecular Glues
2D class averages enhance signal to noise ratio, increasing information that can be gleaned from single particle analysis.
Structural details of individual particles can be difficult to discern from raw images taken on an electron microscope. This is due to the low signal-to-noise ratio typical for images of radiation-sensitive biological samples. It becomes even more challenging when the particles are conformationally or compositionally heterogeneous.
2D class averaging is the first step in visualizing the three dimensional structure of a protein or macromolecule, and can provide researchers a comprehensive representation of particles in their sample. 2D class averages can help visually assess variability in qualities such as preferred orientation, conformation, aggregation, and homogeneity of a sample in order to help select the most viable samples for full 3D structure reconstruction.
To obtain 2D class averages, first we vitrify grids by taking an aliquot of the sample in a buffer and applying it to a grid before plunging it into liquid ethane. The goal is to obtain a thin, uniform layer of ice, with particles embedded in a variety of orientations in order to capture a variety of different views of the sample. By imaging as many different orientations of the sample as possible, we are able to create a more accurate three dimensional reconstruction. 2D classifications can be obtained from both negative stain and cryo-EM micrographs.
In the 2D class averaging process, individual particles are selected from TEM images, aligned relative to each other, and then computationally classified based on apparent similarities. In this way each particle class becomes an average of many similar particles, which enhances the overall signal. These 2D class averages provide a rich source of information on basic particle features, domain structures, and can also help in understanding the degree of particle flexibility.
Our automated methods provide reliable high throughput image acquisition and processing, and help answer questions on critical particle features.